VKORC1 Overview
](https://s3.pgkb.org/pathway/PA145011114.png?versionId=WPnE8UK.Ot6e_jjG2Uq2vvHnlTAiQxNX)
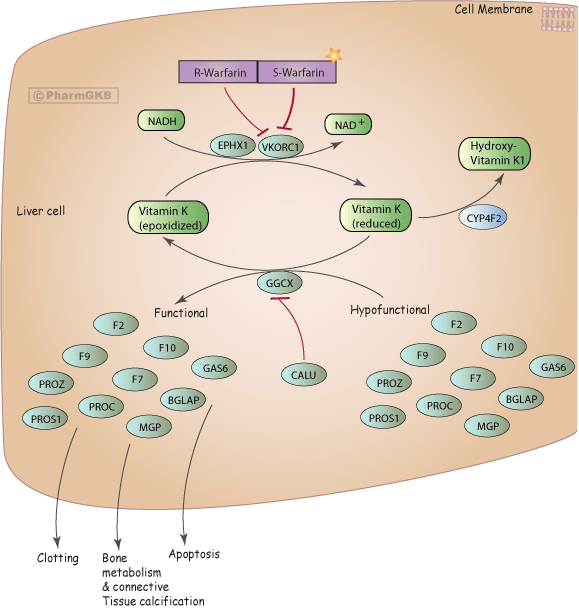
VKORC1 encodes the vitamin K epoxide reductase enzyme, which is responsible for converting vitamin K epoxide to vitamin K as part of the clot signalling pathway. Warfarin is a lifesaving VKORC1 inhibitory medication used to lower the risk of deadly blood clot formation in at risk patient populations, such as the 2.6 million people living with atrial fibrillation in the United States 1. Doctors must carefully prescribe Warfarin: too little and a patient is not treated, too much and the patient could die of internal bleeding. Their jobs are made more difficult by the two key single nucleotide polymorphisms (SNPs) that exist of the VKORC1 gene in the human population, both which affect Warfarin’s ability to bind to the enzyme and inhibit its activity. The first, known as VKORC1A, is caused by a single Guanine to Adenine transversion. This SNP affects the transcription binding site, reducing VKORC1 transcription and consequently increasing Warfarin sensitivity. The second is caused by replacing aspartic acid with tyrosine at position 36, resulting in noticeable Warfarin resistance due to the decreased ability for the enzyme to bind to the drug2. This leads to doses that varying from 2mg to 10mg once per day 3. Locus‐Specific DataBases (LSDB), like the Leiden Open Variation Database (LOVD) and PharmGKB, store information on gene sequence variation associated with human phenotypes and are of particular importance in dosage calculation algorithms and guidelines used by physicians in determining Warfarin dose among diverse patient populations4.
Primary Database: The Leiden Open Variation Database (LOVD)
](https://pbs.twimg.com/profile_images/2455207201/logo_2012-07-20_twitter_400x400.jpg)
The database documentation states that the LOVD, “focuses on the collection and display of DNA sequence variants,” with extensive support for complete clinical data as well as automatic annotation of VCF files. The most recent version 3.0 was released in 2007 and data is still being updated. Full documentation for the Leiden Open Variation Database can be found here.
Types of Data and Biological Questions
The Leiden Open Variation Database can answer many biological questions. As well as including information on the precise chromosomal location of the gene within the human genome, it includes categorized links to the NCBI homepage corresponding to the genomic, transcript, and intron/exon reference for a particular gene. Where this database specializes is in the recording of all publically available variants of a gene from affected individuals, including graphs and statistics on what kinds of mutations occur with what genes, as well as what diseases are commonly associated with what mutations.
Primary Characteristics: Data Collection
The Leiden Open Variation Database is a primary database that mainly collects information on individuals, and variants of genes observed in those individuals, from registered submitters. All submitters must go through a vetting process whereby their academic credentials and institutional associations are reviewed by curators and managers of the service. A complete submission contains information on an individual, disease and phenotype information (optional), variant screening(s), and at least one found variant. There a curator reviews the data and publishes the acceptable entries on the main webpage on the behalf of the submitter. Managers oversee curators and are responsible for large scale manipulations to the database.
Data Storage, Format, and Usage
Since the Leiden Open Variation Database is most concerned with the collection of genomic variants, it mainly accepts VCF, or Variant Call Format files, a type of Next Generation Sequencing format. The database is divided into five categories: genes, transcripts, variants, individuals, and diseases. Searching the LOVD is extensive, with twenty-five categories under “Variants” alone. Some searchable categories include haplotype, the database ID, the genomic DNA change that causes a particular variant, and remarks made by the curator who submitted the data. While the LOVD includes many categories of information, some subcategories like “Published as” and “Frequency” seem seldom used. These and many other subcategories are sometimes left blank, providing a patchwork array of varying information that can be difficult to navigate. Well documented code extensions as well as “out of the box” design functionality work to help make the Leiden Open Variation Database accessible to as many users as possible.
Restrictions
The front end web interface of the Leiden Open Variation Database is open source and freely viewable to the public. However, the contents of this LOVD database are the intellectual property of the respective curators and are protected by copyright. Hidden from public view is the data input by registered submitters which has not been published by a curator or a manager. This is either because the data has not yet been processed, the data is not yet up to the standards of the database management team, or the data is private and will never be shared publicly. Numerous security measures are in place to keep user accounts and their associated data secure, including a safe login procedure, resistance to SQL injection and cross-site scripting, as well as password protected data manipulation forms. What is visible to the public is just the tip of the iceberg in terms of what data might be stored in the database.
LOVD in Literature
The Leiden Open Variation Database is a lesser known yet extensively documented database. The following review article describes the LOVD’s open source, “out of the box” design philosophy for easy maintenance and management: LOVD v.2.0: the next generation in gene variant databases.
VKORC1 in LOVD
A search for VKORC1 in the LOVD returns the gene homepage, with basic information about the gene automatically curated by NCBI. Available are an informative series of graphs that explain the types and proportions of genetic polymorphisms for the gene. From this view it is easy to see that the database contains 187 public variant entries, with 20 unique variations, 90% of which are substitutions.
Secondary Database: PharmGKB
The process by which clinicians determine guidelines for drugs like Warfarin is called Pharmacogenomics, the study of the relationship between genetic variations and how the human responds to medications. PharmGKB is a secondary database that serves to curate knowledge about the impact of genetic variation on drug response, serving as a resource to both researchers and physicians alike5.
Types of Data and Biological Questions
PharmGKB works mainly to store information which supports the reasoning behind the clinical guidelines which determine the dosage requirements of various medications. In addition to listing common gene variants as in the Leiden Open Variation Database, PharmGKB works to help clinicians interconnect the abstract study of genomics with the practice of clinical medicine. For example, an oncologist may wish to understand the pharmacogenetics of certain cancers in order to better understand what drugs to use to treat them. Clinicians can also access primary literature pertinent to the gene, gene variant, or drug of study, and can even find similar molecules which may work as experimental alternatives.
Secondary Characteristics: Data Collection
PharmGKB is a secondary database which uses a combination of natural language processing methods, as well as hierarchical, in-house manual curation to mine clinical reports, journal articles, as well as primary databases to provide high quality pharmacogenomic data. Every entry includes a links and downloads section to help users visit the automatically generated portions of the database.
Data Storage, Format, and Usage
](https://3.bp.blogspot.com/-rggZzD7zCTw/VgQdIK7gChI/AAAAAAAABt4/tA5WoOePKXU/s1600/pyramid-brokenout-web.png)
PharmGKB is remarkable for its streamlined look and ample use of formatting on its webpage, creating an experience that is removed from typical databases. For example, Variant Annotation tables for particular genes include links to publications. The search bar suggests users add search terms together to create new combinations, in order to discover compare and contrast drugs, genes, and more. Format is in the form of lists of annotations and links to publications, with many sources pointing back to “AmiGO 2,” a primary gene ontology database. The above image outlines how PharmGKB curates and selects it’s content. The data found on PharmGKB is mainly used to determine drug interactions, dosing guidelines, as well as genes of pharmacogenomic significance for clinical researchers.
Restrictions
PharmGKB is managed by Stanford University and is published under a Creative Commons Attribution-ShareALike 4.0 license. This public license allows users to freely access and share the information found in PharmGKB.
PharmGKB in Literature
The following article titled VKORC1 Pharmacogenomics Summary uses PharmGKB as it’s primary database reference to study the clinical affects of three different VKORC1 variants. Additional literature sources automatically curated by PharmGKB can be found here.
VKORC1 in PharmGKB
A search of VKORC1 in PharmGKB returns 47 instances of prescribing information, 33 clinical annotations, and one mechanism of action. VKORC1 is also listed as a very important pharmacogenetics gene summary due to its stringent dosing requirements highly dependent on genetic polymorphisms. Additional information consists of location information as well as enzyme and gene names. VKORC1 is located on chromosome 16, “GRCh38.p7 - chr16 : 31090842 - 31094999.” 6 Along with common gene information, PharmGKB includes a large number of prescription study annotations that serve to support the five different sets of clinical guidelines put forth for VKORC1 and other similar genes that affect Warfarin sensitivity.
Comparison and Conclusion
Locus‐Specific Data Bases (LSDB), like the Leiden Open Variation Database (LOVD) and PharmGKB, play similar roles in annotating variants of genes such as VKORC1. However, unlike LOVD not just anyone can contribute to PharmGKB. Beacause it features secondary characteristics like dosing instructions intended for use by physicians in real world settings, the curators of PharmGKB justifiably choose not to accept primary information submitted through the website, but rather primary information mined from databases of journal articles which have undergone extensive peer review. While both include detailed curation by human experts, PharmGKB employs natural language processing techniques to help assemble as much information as possible. For all the computational tools employed by PharmGKB curators, the LOVD includes detailed and extensive documentation that dives into the structure of the database, code extensions, and curation procedures used to create and manage the database. This ease of management comes at the cost of transparency; LOVD is challenged by an outdated user interface and somehwhat awkward controls. Overall these databases differ in ther primary and secondary characteristics. What makes the Leiden Open Variation Database a primary database is it’s emphasis on manually uploaded data from individual researchers in small scale clinical settings, while PharmGKB is defined by it’s agglomeration of clinical trials and individual studies pubished elsewhere in other primary databases as well as medical journals.
-
What is Atrial Fibrillation (AFib or AF)? (n.d.). Retrieved October 15, 2019, from www.heart.org website: https://www.heart.org/en/health-topics/atrial-fibrillation/what-is-atrial-fibrillation-afib-or-af ↩
-
VKORC1 gene homepage—Global Variome shared LOVD. (n.d.). Retrieved September 28, 2019, from https://databases.lovd.nl/shared/genes/VKORC1 ↩
-
Owen, R. P., Gong, L., Sagreiya, H., Klein, T. E., & Altman, R. B. (2010). VKORC1 Pharmacogenomics Summary. Pharmacogenetics and Genomics, 20(10), 642–644. https://doi.org/10.1097/FPC.0b013e32833433b6 ↩
-
Fokkema, I. F. A. C., Taschner, P. E. M., Schaafsma, G. C. P., Celli, J., Laros, J. F. J., & Dunnen, J. T. den. (2011). LOVD v.2.0: The next generation in gene variant databases. Human Mutation, 32(5), 557–563. https://doi.org/10.1002/humu.21438 ↩
-
M. Whirl-Carrillo, E.M. McDonagh, J. M. Hebert, L. Gong, K. Sangkuhl, C.F. Thorn, R.B. Altman and T.E. Klein. “Pharmacogenomics Knowledge for Personalized Medicine” Clinical Pharmacology & Therapeutics (2012) 92(4): 414-417. ↩
-
VKORC1—Overview. (n.d.). Retrieved October 15, 2019, from PharmGKB website: https://www.pharmgkb.org/gene/PA133787052/overview ↩